Search tips for finding gene pages
Here are some search tips if you are having trouble finding genes you are looking for. The two most likely problems are:
2) My search returns too many genes
1) I can't find my gene
As most genes are being renamed to align S. purpuratus with the human genome nomenclature committee (HGNC) names, searching by an old name may not work. We do store alternative gene names as synonyms in the database so if it is there, your search will find it. If it fails it means we don't yet have that name in the database. Try the human, mouse or zebrafish gene name if you know them. If this fails also, the ultimate gene finder is blasting the Echinobase database.

Select blast flavor and target (protein or mRNA) and click the "Submit Job" button.
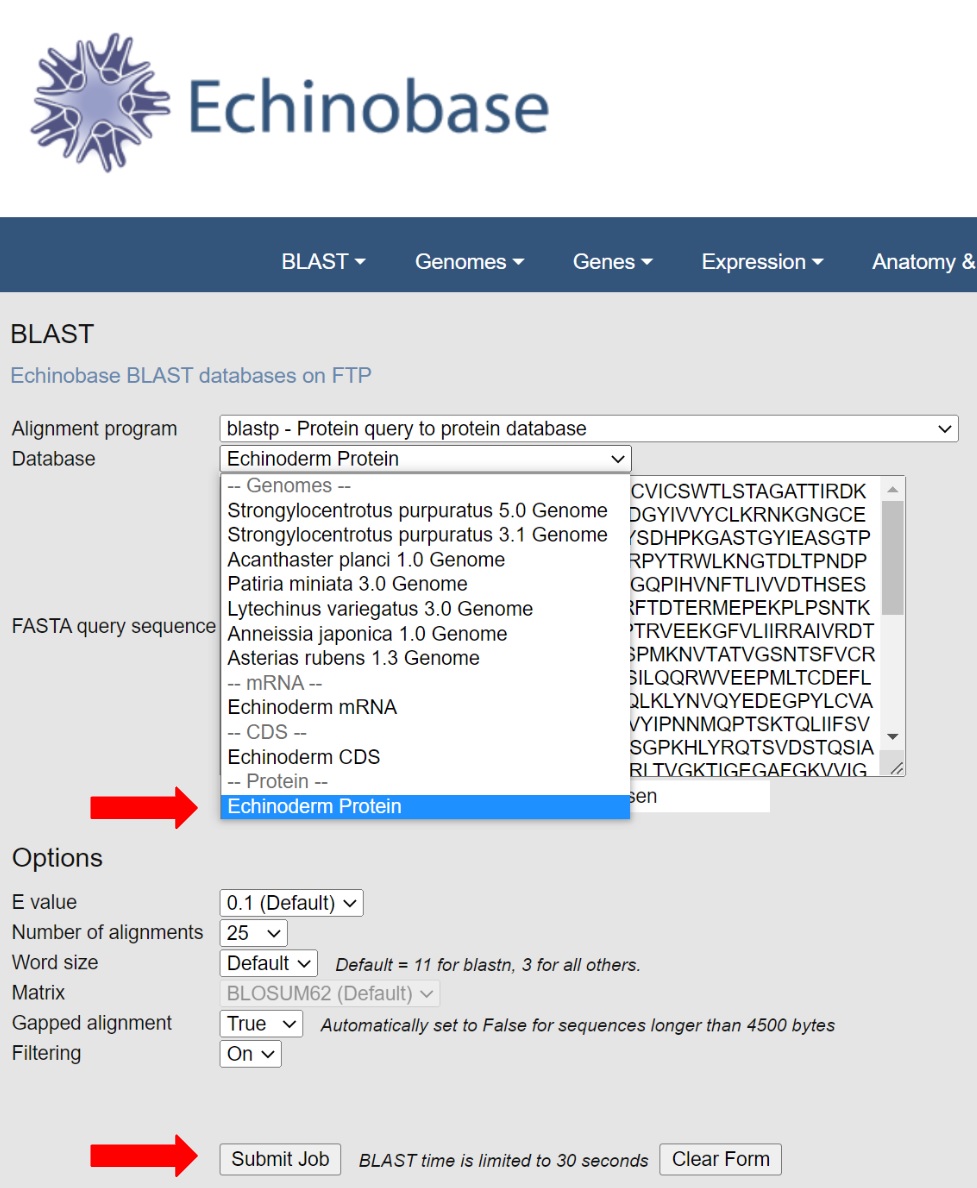
Blast returns have links to matching gene pages so you can jump straight to your gene by simply clicking on the gene name link in the result.
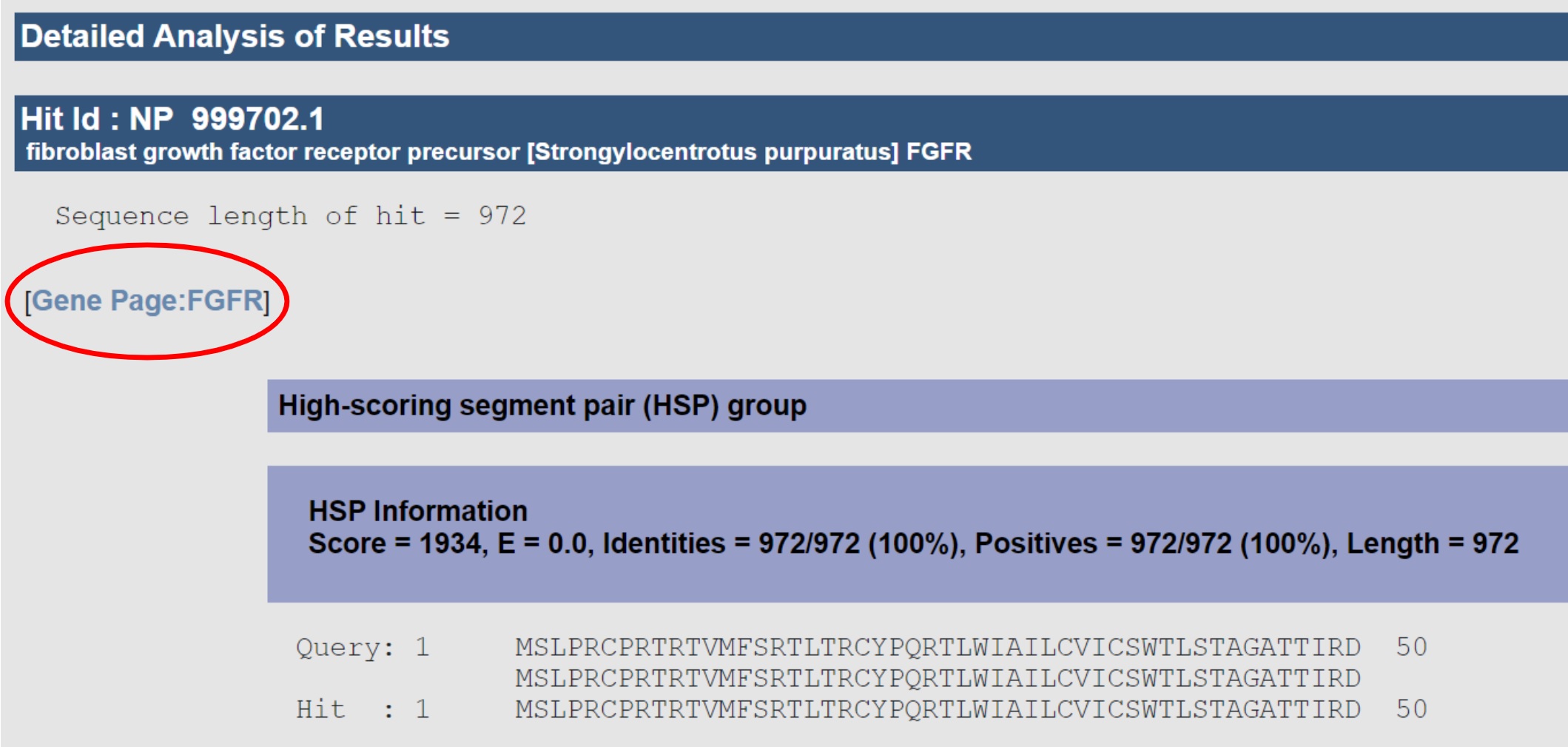
If you find a gene page matching an old gene name, and that name is not displayed as a synonym at the top of the page, you could do us and your colleagues a great favor by adding it--then next time somebody searches for that name they’ll find it. We also use synonyms to link to paper titles and abstracts and to figure legends, so adding one small synonym can result in a plethora of links to extra information. To add a synonym you need a Echinobase account, then select the link right next to the synonym field.
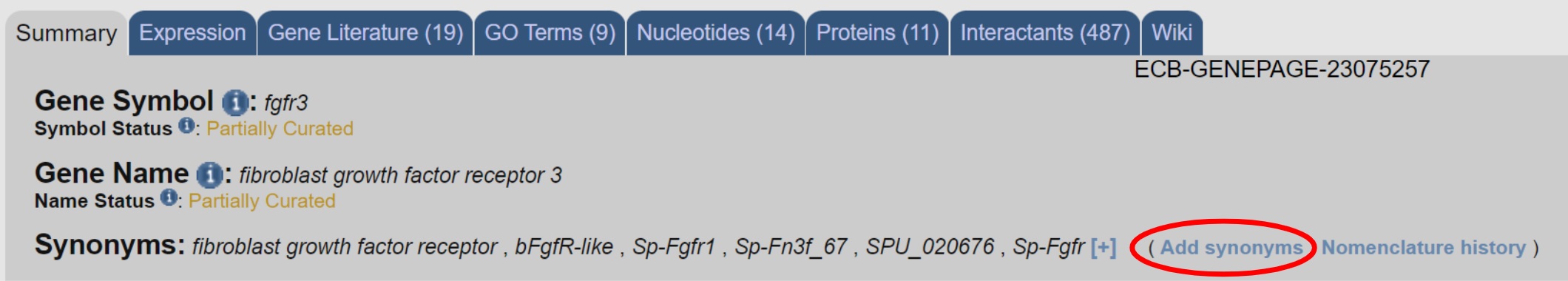
Note how this page has 5 associated publications. Some were linked via synonyms and not via the new official name, fgfr. It will also allow us to link to your personal Echinobase page.
2) My search returns too many genes
The search function is set to search all possible fields as the default. This means it looks in names, synonyms, orthologs--everywhere--which is why lots of genes can show up. Your search phrase is highlighted in the search returns to help you see why your search is finding so much stuff.
To find fewer hits try using a more selective search term using the drop-down menu.
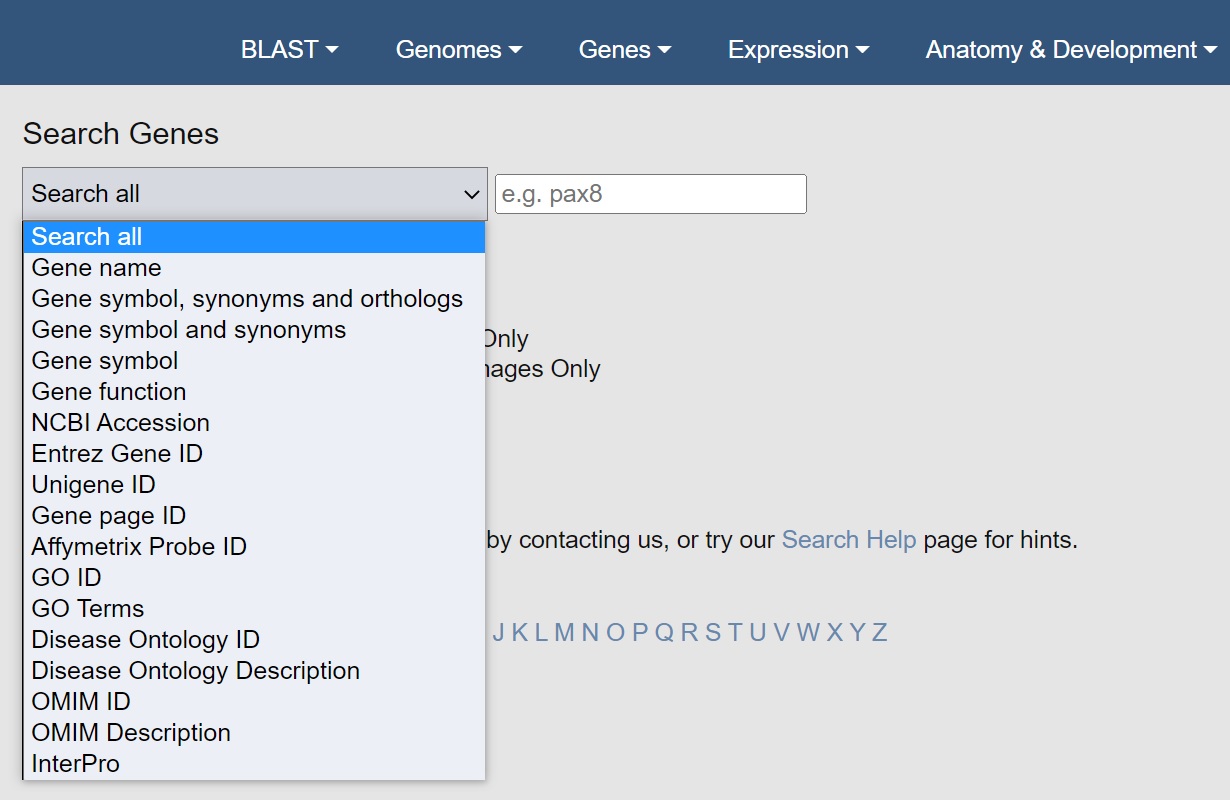
We suggest you try "Gene symbol and synonyms" first. This will only search gene names, both new HGNC style names and old legacy names (if we have them). It usually brings down the number to a reasonable scale. If it is still too many hits try selecting "Gene symbol", which will now miss synonyms but will be nice and restricted.
