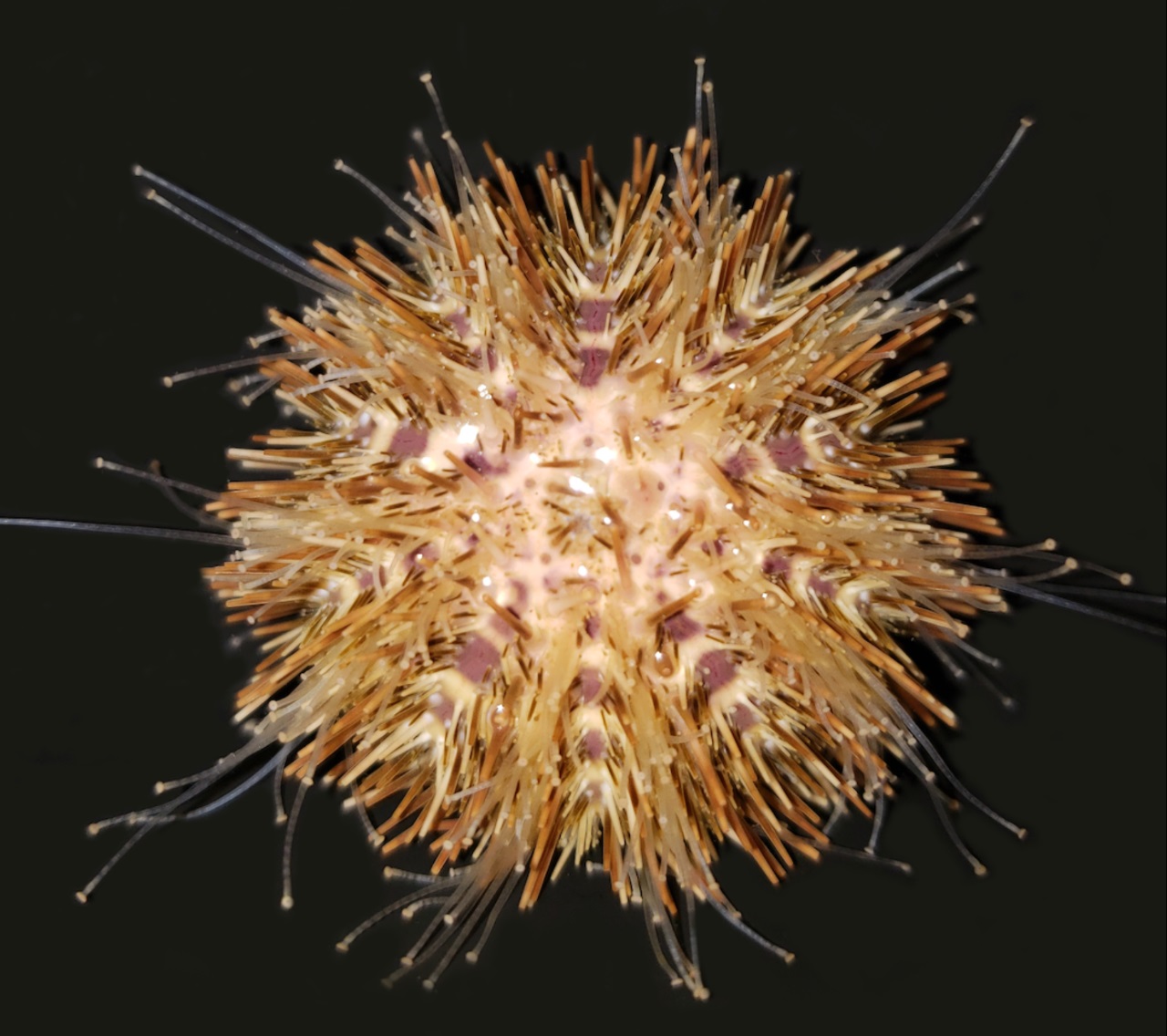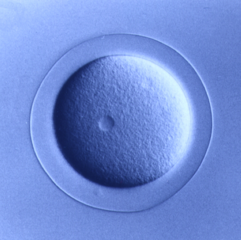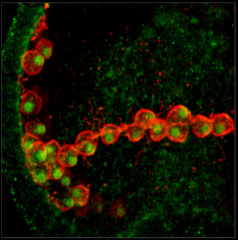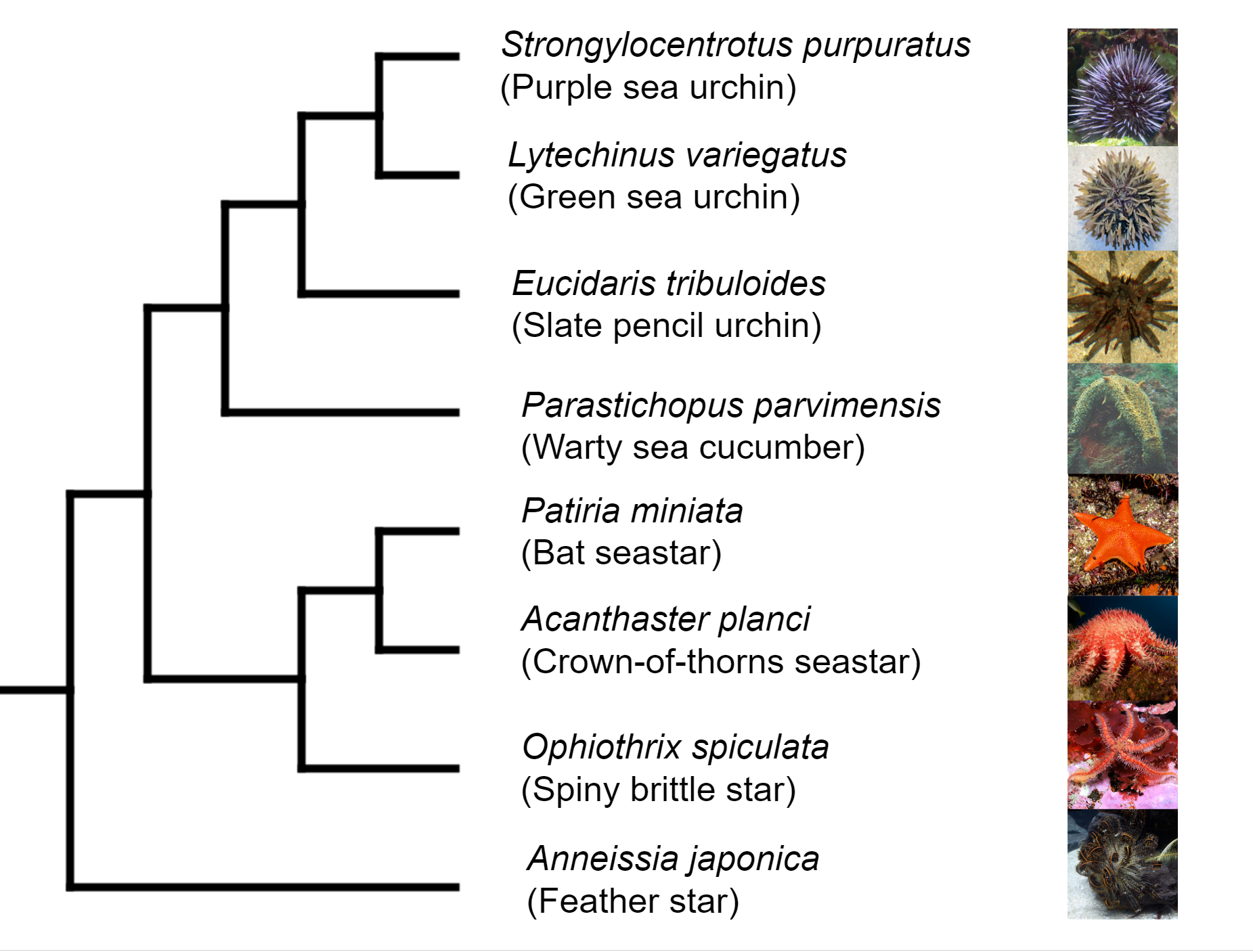
Deuterostomia (superphylum) includes Chordata, Echinodermata and
Hemichordata phylum, Echinodermata emerged in the Lower Cambrian
(> 540 million years ago, MYA) and split from Chordata 400-500
MYA, Echinodermata includes 7,000 extant and 13,000 extinct,
marine only species. Echinodermata has 5 Classes: the basal
branching Crinoidea (crinoids), and the 4 motile Eleutherozoa
Classes including Asterozoa, the Ophiuroidea (brittle stars) and
Asteroidea (starfish) and the Echinozoa, the Echinoidea (sea
urchins and sand dollars) and Holotheroidea (sea cucumber).
Echinoderms are
model organisms for studying embryo development
and regeneration due to many unique features including:
- the ability to synchronize fertilization of millions of
eggs
- transparent embryos and larvae
- varied development within and between a genus
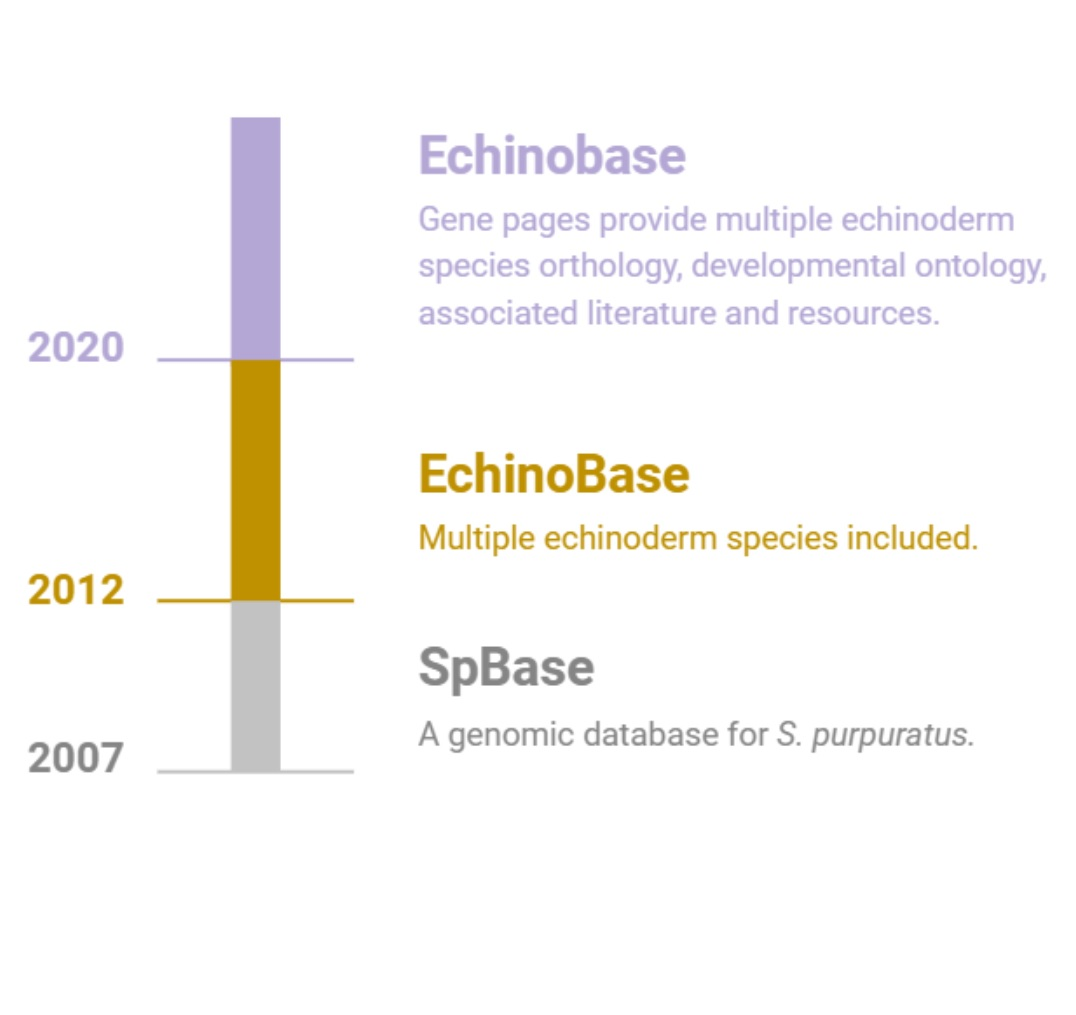
Echinobase supports the international research community by providing a centralized, integrated and easy to use web based
resource to access the diverse and rich, functional genomics data of echinoderm species.
Echinobase is organized around the GENEPAGE which displays information about genes, orthology, and links to research papers. The gene
models of echinoderm species are associated with the genome sequence using the genome browser, JBrowse. Temporal developmental gene expression
data will be displayed when available, and spatial cell type information will be associated using the Echinoderm Anatomy and Development Ontology (ECAO).
Echinobase provides a critical data sharing infrastructure for other NIH-funded projects. In addition to our primary goal of supporting echinoderm
researchers, Echinobase enhances the availability and visibility of echinoderm data to the broader biomedical research community.
Species in Echinobase are divided into two categories:
fully supported species found on gene pages and
partially supported species that only have support through the genome browser and BLAST processes. We are an expanding resource and both species sets will grow in upcoming months and years.
More About Echinobase

S. purpuratus
Strongylocentrotus purpuratus (the purple sea urchin)
The purple sea urchin,
Strongylocentrotus
purpuratus, lives in lower intertidal and nearshore subtidal
communities along the eastern edge of the Pacific Ocean extending
from Ensenada, Mexico to British Columbia, Canada. Morphological
development of
S. purpuratus from
embryo to adult has been studied for over 100 years and recent
decades have added genetic and genomic data to our knowledge of
this model organism.
The substantially improved current release of the purple
sea urchin genome assembly (Spur5.0) is sixth in the series.
Learn more about the
genomics of the purple sea urchin.
Lytechinus pictus (painted urchin)
Lytechinus pictus, the painted urchin, lives in the shallow reefs off the Pacific coasts of California and Mexico.
This sea urchin is small, only 40 mm (1.6 in) in diameter. The general color is white or pale straw brown, sometimes pinkish.
L. pictus is being developed as a
genetically-enabled sea urchin model. Genetic lines of
L. pictus have been generated with stable modifications that are useful for cell and developmental studies as
well as physiological studies and drug screens.

A. japonica
Anneissia japonica (feather star)
The crinoids are sessile and include feather stars, which do not have a stalk, and sea lilies which are attached to the sea floor by a stalk. Feather stars are most abundant in shallow, rocky waters of the Indian and western Pacific oceans. As plankton flows by the feather-like arms it sticks to a mucus secreted by the tube feet and these particles are moved towards the mouth.
Species of crinoids are used as an outgroup for phylogenetic studies within the Echinodermata.

Ptychodera flava
Ptychodera flava (Yellow acorn worm)
Ptychodera flava, commonly known as the yellow or Hawaiian acorn worm, is a member of the phylum Hemichordata and is widely distributed across marine ecoregions throughout the Indo-Pacific. Like other acorn worms,
P. flava has a distinctive three-part body plan consisting of a short, acorn-shaped proboscis, a collar region, and a long posterior trunk. It typically lives buried beneath the sand, using its proboscis to sift and forage for food.
Hemichordates are generally regarded as the sister group to echinoderms and occupy an important evolutionary position between vertebrates and invertebrates. This makes
P. flava a valuable model organism for studying the genomic and evolutionary origins of chordate and deuterostome traits.

A. filiformis
Amphiura filiformis (Brittle star)
Amphiura filiformis (O. F. Muller, 1776) is a brittle star found on the seabed of the European coasts from Greece to Norway and Sweden including the Baltic Sea. It is an important member of soft bottom communities where it digs a shallow burrow and feeds on plankton. It serves as food for crayfish and finfish.
Stemming from its remarkable ability to regenerate arms in a matter of weeks it has become an emerging model for regeneration and stem cell biology in biomedical research. Research targets include:
- The cellular and molecular mechanisms underlying regeneration with links to neuroscience, stem cell biology and neuropeptide structure and function in the absence of a centralised nervous system.
- Comparative developmental events involving the evolution of biomineralization in the echinoderm clade.
- Support for phylogenomic analysis in an otherwise little studied group.
Development raw reads are available at NCBI accession
PRJNA349786.

A. rubens
Asterias rubens (sugar star or the common sea star)
The sugar star is the most familiar and common sea star in the northern Atlantic coastal waters, living on rocky and gravelly surfaces. The five arms are orange or brownish in color and span 10-30 cm. Sugar stars feed on molluscs, polychaete worms and barnacles and can be preyed upon by the common sunstar.

P. miniata
Patiria miniata (the bat star)
The bat star,
Patiria miniata,
(formerly called
Asterina miniata)
is a common sea star found along the Pacific coastline of North
America from southern Alaska to the Gulf of California, Mexico,
in intertidal and subtidal locations to depths of 300 m. Bat stars
typically have 5 arms but range from 3 to 8 and can be red,
orange, brown, yellow, blue, pink, green, and intermediate color
and adults are about 8 inches across.
P. miniata reproduce by
broadcast spawning and embryos hatch into planktonic larvae and
later metamorphose into pentaradial juveniles which develop into
young sea stars with stubby arms.
The current version, v3.0, of the assembly is fully
supported at Echinobase.
Learn more about the
genomics of the bat star.

L. variegatus
Lytechinus variegatus (the green sea urchin)
Lytechinus variegatus (Lamarck, 1816) is known as the green sea urchin or
variegated sea urchin. It has many color morphs including white, green, purple, red, or pink or a
combination of those colors. Multiple subspecies inhabit distinct geographical territories of the
western Atlantic tropics ranging from North Carolina south to the Carribean, Gulf of Mexico and
northern Brazil. The planktonic larvae are known as pluteus larvae that metamorphose to juvenile
urchins.
The
L. variegatus v3.0 genome is the first echinoderm assembly with
chromosomal resolution.
Learn more about the
genomics of the green sea urchin.

A. planci
Acanthaster planci (the crown-of-thorns seastar)
The crown-of-thorns seastar (COTS),
Acanthaster
planci, is is named for the spines that cover the top of its
large body and multiple arms.
A.
planci lives in Indo-Pacific waters and preys upon corals.
Outbreaks where large numbers of COTS aggregate can decimate
coral reefs. Due to their destructive nature the genome was
sequenced in the search for biocontrol strategies. The current
version of the
A. planci genome,
v1.0, is available on Echinobase.
Learn more about the
genomics of the crown-of-thorns seastar.
Echinoderms are marine organisms that when harvested from the
wild, can live in aquariums and embryo and larval development has
been studied for over 100 years. Echinoderm eggs are fertilized
externally and embryos and larvae are transparent, enabling
detailed descriptions of the structure and timing of development.
The variation and comparison of developmental patterns of
related species is used to improve understanding of these vital
processes and their evolution in echinoderms and deuterostomes.
Echinoderms are models for studies of skeletogenesis and
regeneration.
The Echinoderm
Anatomical Ontology (ECAO) provides standardized terms for
developmental stages for S.
purpuratus, and is being expanded to include additional
echinoderm species.
For studies of regulatory sequences BAC Libraries are available.
Refer to the
BAC
Table for those that can been identified using BAC-ENDs and the
genome browser or screen the BAC library for your gene of
interest. An in-frame fusion of a reporter gene can be engineered
into the locus of interest by recombineering to assess gene
expression and putative regulatory sequences.
Echinobase
automatically searches the scientific literature
and collects articles related to echinoderms. Users can
come to the site to access currently available published research
in one
centralized location. Papers are
manually
curated to link genomic information to protocols and reagents for
future studies.
 Deuterostomia (superphylum) includes Chordata, Echinodermata and
Hemichordata phylum, Echinodermata emerged in the Lower Cambrian
(> 540 million years ago, MYA) and split from Chordata 400-500
MYA, Echinodermata includes 7,000 extant and 13,000 extinct,
marine only species. Echinodermata has 5 Classes: the basal
branching Crinoidea (crinoids), and the 4 motile Eleutherozoa
Classes including Asterozoa, the Ophiuroidea (brittle stars) and
Asteroidea (starfish) and the Echinozoa, the Echinoidea (sea
urchins and sand dollars) and Holotheroidea (sea cucumber).
Deuterostomia (superphylum) includes Chordata, Echinodermata and
Hemichordata phylum, Echinodermata emerged in the Lower Cambrian
(> 540 million years ago, MYA) and split from Chordata 400-500
MYA, Echinodermata includes 7,000 extant and 13,000 extinct,
marine only species. Echinodermata has 5 Classes: the basal
branching Crinoidea (crinoids), and the 4 motile Eleutherozoa
Classes including Asterozoa, the Ophiuroidea (brittle stars) and
Asteroidea (starfish) and the Echinozoa, the Echinoidea (sea
urchins and sand dollars) and Holotheroidea (sea cucumber).  Echinobase supports the international research community by providing a centralized, integrated and easy to use web based
resource to access the diverse and rich, functional genomics data of echinoderm species.
Echinobase supports the international research community by providing a centralized, integrated and easy to use web based
resource to access the diverse and rich, functional genomics data of echinoderm species.